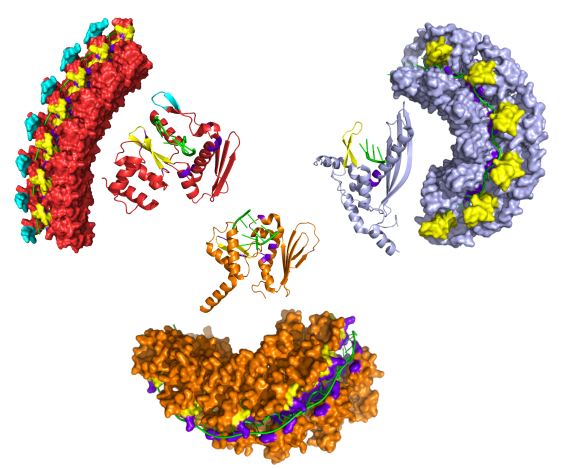
A structural phylogenetic tree of Rad52 and its annealase superfamily
- Principal Investigators:
- Prof. Dr. Michael Schroeder
- Project Manager:
- Ali Al-Fatlawi
- HPC Platform used:
- NHR@TUD Barnard + Taurus
- Project ID:
- p_drug
- Date published:
- Introduction:
- Proteins are the workhorses of cells, driving all processes essential for survival. Each protein folds into a specific shape, uniquely tailored to its sequence and function. Predicting protein structures from sequences has long been one of biology’s greatest challenges. Recent breakthroughs in computational methods, like AlphaFold, which earned the 2024 Nobel Prize in Chemistry, have revolutionized this field.
- Body:
-
The Schroeder group specializes in computational techniques to analyse protein structures and sequences, uncovering evolutionary relationships that can inform drug development and disease diagnosis. A key focus is on DNA repair proteins such as human Rad52 and bacterial RecT and Redβ. Despite their vastly different sequences, these proteins share a very similar structure.
Rad52 is a cancer target involved in DNA repair and genomic stability. Investigating the relationship between human Rad52 and prokaryotic DNA repair proteins can enhance our understanding of cellular mechanisms and functions. With the recent advent of AI-based protein structure prediction, we can now observe evolutionary relationships at an unprecedented level of detail. This project explores these new possibilities by conducting the largest structural analysis to date of 10,000 proteins in the Rad52 superfamily, which is classified into five families using advanced sequence-based methods. We examine these proteins from a structural perspective and construct a phylogenetic tree based on predicted structures, uncovering intricate relationships that shed light on how these proteins are related, particularly between human and bacterial proteins. While sequence-based methods often struggle to detect distant homologs due to high divergence, structural analysis reveals conserved folds, enabling more accurate predictions of functional relationships between SSAPs across species.Publications
1- Al-Fatlawi, A., Menzel, M. & Schroeder, M. Is Protein BLAST a thing of the past?. Nature Commun 14, 8195 (2023).
2- Al-Fatlawi, A., Schroeder, M. & Stewart, A.F. The Rad52 SSAP superfamily and new insight into homologous recombination. Commun Biol 6, 87 (2023).
3- Al-Fatlawi A., Md Hossen, B., de Paula Lopes, S., Stewart, F., Schroeder M., A structural phylogenetic tree of Rad52 and its annealase superfamily, Computational and Structural Biotechnology Journal, accepted paper (2024).
4- Al-Fatlawi A., Md Hossen, B., El-hendi F., Schroeder M. "Protein secondary structure and remote homology detection." bioRxiv (2024).
- Institute / Institutes:
- Biotechnology Center (BIOTEC), Center for Molecular and Cellular Bioengineering
- Affiliation:
- Technische Universität Dresden
- Image:
-