Bridging Researchers and High-Performance Computing for Advanced Bio-Image Analysis
- Principal Investigators:
- Dr. Robert Haase
- Project Manager:
- Dr. Otger Campàs
- additional Affiliation:
- Center for Scalable Data Analytics and Artificial Intelligence (ScaDS.AI); Helmholtz AI, Helmholtz Zentrum Dresden Rossendorf
- HPC Platform used:
- NHR@TUD Taurus (alpha)
- Project ID:
- p_bioimage
- Date published:
- Researchers:
- Dr. Till Körten, Dr. Johannes R. Soltwedel, Dr. Marcelo L. Zoccoler
- Introduction:
-
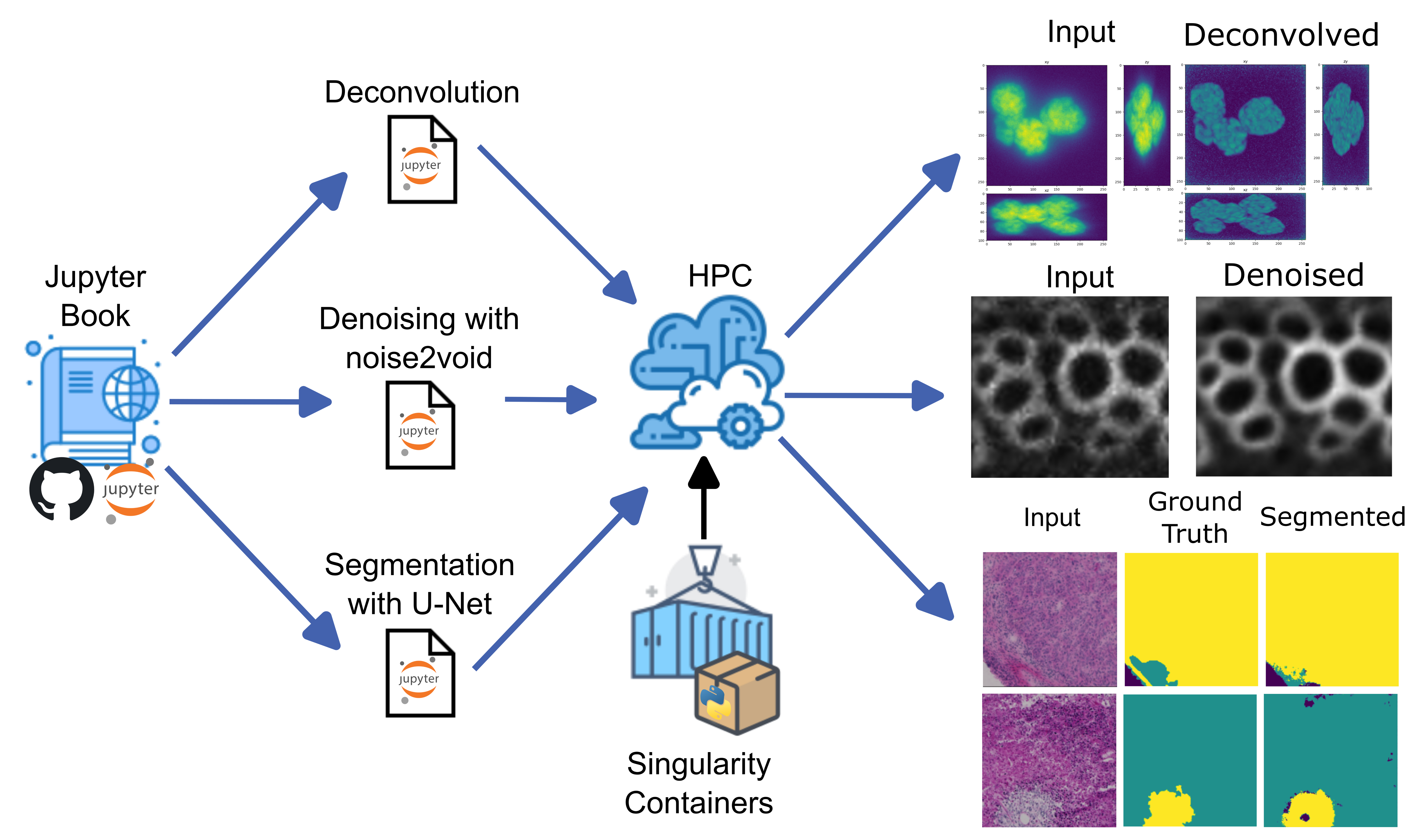
We aim to make sophisticated, computationally demanding bio-image analysis workflows more accessible to researchers who have little or no programming background. Thus, we organized a course that leveraged the Jupyter Hub interface on an HPC cluster in conjunction with custom Singularity containers, each tailored to the specific needs of individual projects. This setup enabled seamless execution of tasks such as three-dimensional image deconvolution, deep- learning–based segmentation (U-Net), and denoising algorithms, all within a user-friendly environment.
Participants in the course successfully ran these resource-intensive workflows without needing to install or configure complex dependencies. These results demonstrate the reproducibility and
efficiency of our container-based approach, which significantly lowers the barrier to adopting HPC cluster solutions for bio-image analysis. Ongoing efforts include expanding our collection of containers to cover a broader range of tasks, integrating OMERO for enhanced data management, and investigating large language model (LLM)–assisted container creation to further automate and streamline workflow setup.
- Body:
-
Introduction
Quantitative bio-image analysis is both a continuously expanding field of research and an increasingly demanding service. As modern high-resolution microscopes typically produce three- dimensional datasets on the order of gigabytes, data analysis cannot be performed on local individual computers anymore. Besides their size, these data are usually complex and may require sophisticated, computationally consuming, deep-learning algorithms for analysis. High- performance computing (HPC) clusters address these needs by providing scalable and cost- effective data processing.
Is spite of these capabilities, many researchers remain unaware of available HPC resources and struggle to manage vast datasets. Introductory courses on HPC usage are available, however some field-specific operations, like environment setup and job submission for image analysis, can still discourage its usage. Leveraging HPC clusters accelerates data analysis, reduces costs, and offers substantial storage capacity, making them highly advantageous for scientists handling large-scale computational tasks. The primary goal of this project was to train researchers with limited programming expertise that work with image data on how to best utilize on-campus resources to process their microscopy images effectively. Therefore, we developed an environment setup workflow and organized a training school to teach researchers how to best utilize HPC clusters for complex large bio-image data analysis.
Method
We have established a workflow that creates singularity containers [1] with a specific set of Python packages targeted for different bio-image analysis workflows. This strategy allowed us to enable these environments as Jupyter kernels so that the user could, from the HPC Jupyter Hub interface, select these kernels and have the necessary packages and correct versions readily
available to run the code. As a way to advertise and distribute this functionality, we organized a course called “PoL Bio-Image Analysis Training School on GPU-Accelerated Image Analysis” [2] where we taught local and international researchers how to run deconvolution in 3D images, how to train a U-Net model for segmentation and how to run a denoising deep-learning algorithm using the HPC cluster.
Results
The participants of the course were able to smoothly run the Jupyter notebooks in the cluster and complete the proposed bio-image analysis tasks. This achievement demonstrated the reproducibility of the proposed workflow and empowered these researchers with an easier way to run bio-image analysis workflows that are computationally demanding and that would otherwise be very hard to set up manually in the cluster or in individual workstations.
Ongoing work/outlook
We are currently working on providing more containers targeted at specific bio-image analysis tasks, but also tailored containers that combine multiple tasks for collaborators that need to orchestrate multiple tasks in a single workflow. The Jupyter notebooks needed for running these workflows could be built with the help of bio-image analyst experts, such as members of our group, or by using available notebooks produced by the community [3].
We are also working on implementing proper data management by integrating data transfer with an OMERO server [4,5]. Besides, we aim to add large language models (LLM) to the singularity container building process in order to build a container based on a given example workflow that is able to identify the necessary packages for the container and that can edit the workflow to redirect image paths to the right places in OMERO, making the environment setting seemingly inconspicuous.
References[1] Sylabs Inc & Project Contributors. “SingularityCE User Guide”, sylabs, 21 Jan 2025,
https://docs.sylabs.io/guides/latest/user-guide/index.html
[2] S. Rigaud, B. Northan, T. Korten, N. Jurenaite, A. Deepak Kulkarni, P. Steinbach, S. Starke, J. Soltwedel, M. Albert, R. Haase. ‘’PoL Bio-Image Analysis Training School on GPU-Accelerated Image Analysis”, biapol.github, 5 Jun 2024, https://biapol.github.io/PoL-BioImage-Analysis-TS- GPU-Accelerated-Image-Analysis/intro.html[3] von Chamier, L., Laine, R.F., Jukkala, J. et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat Commun 12, 2276 (2021). https://doi.org/10.1038/s41467-021-22518-0
[4] Allan, C., Burel, JM., Moore, J. et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods 9, 245–253 (2012). https://doi.org/10.1038/nmeth.1896
[5] T.T.Luik. “BIOMERO - BioImage analysis in OMERO”, nl-bioimaging.github, 21 Jan 2025, https://nl-bioimaging.github.io/biomero/readme_link.html
- Institute / Institutes:
- Physics of Life (PoL)
- Affiliation:
- Technische Universität Dresden
- Image:
-